Recent studies show that human gut microbiota plays a key role in regulating human health. Yet despite this, there are no clinical diagnostic tools that use microbiota to identify imbalances within our gut that can lead to serious disease, or tools for identifying existing disease.
Human microbiota consist of various microbial communities that live in and on our bodies. These communities are composed of different species of bacteria, archaea, protists, fungi and viruses. Together, they constitute complex and diverse ecosystems that interact with us as their human hosts. When we refer to microbiota with their genomic information, we use the term microbiome.
Previous research estimates that the genes in our microbiome outnumber human genes by two orders of magnitude. As the host of microbiota, their presence benefits our bodies by facilitating important chemical processes such as maintaining homeostasis, developing our immune systems and helping our bodies harvest various nutrients that are otherwise inaccessible.
Research also reports that altered states of microbiota within our bodies can contribute to cancer and can adversely affect therapeutic responses in cancer patients. Additionally, several studies have reviewed the role of microbiota in human health and disease, recommending their use for disease diagnosis and prediction.
Microbiome in the age of big data
Over the past two decades, several large-scale, microbial profiling projects were established including the Human Microbiome Project and MetaHIT (Metagenomics of the Human Intestinal Tract) Project. These projects aim at investigating microbial components of the human genetic and metabolic landscape and their link to diseases.
Despite various attempts to develop unified best practices, truly standardized approaches in microbiome research have not yet been established. Missing are statistical and machine learning models that leverage high-throughput, metagenomic data with supervised and unsupervised learning techniques. Two novel, machine learning techniques that are gaining acceptance within microbiome research are Shotgun metagenomic sequencing and 16S rRNA gene sequencing technology. Shotgun metagenomic sequencing allows comprehensive sampling of all genes in microorganisms that are present in a given sample. This technology enables researchers to examine microbial diversity and detect the abundance of genes in different environments. In comparison to 16S rRNA gene sequencing technology, shotgun metagenomic sequencing provides higher resolution of marker profiles at relative species and strain levels.
Existing machine learning methods that examine the gut health of subjects use shotgun metagenomics to differentiate healthy people from those at risk or suffering from disease. They achieve this by extracting and analyzing gut microbial, relative-species abundance and strain-level marker profiles. Both of these gut microbial features show diagnostic potential and have been used separately in previous research for microbiome-based disease prediction.
In addition to relative species-abundance and strain-level marker profiles, metabolite profiles can provide information about the metabolism of gut microbiome. By combining metabolomic and metagenomic data, we can get a more complete description of an individual’s heath. Metabolomic data can be extracted for analysis using different techniques such as capillary electrophoresis, time-of-flight mass spectrometry (CE-TOFMS).
Advancing disease prevention and treatments with NEC MicrobiomePredict for microbiome-based disease prediction
At NEC Laboratories Europe, we have proposed the machine learning model and system, NEC MicrobiomePredict. The system uses Multimodal Variational Information Bottleneck to analyze a person’s gut microbiome and predict whether they are suffering from a disease (see Figure 1).
MicrobiomePredict is a microbiome-disease classification method; it leverages the theory of Information Bottleneck (IB) to learn joint encoding from different input data modalities such as relative species-abundance, strain-level marker profiles and metabolomic data. The joint encoding learned by MicrobiomePredict applies maximum compression to heterogeneous data modalities that are input, while preserving all the information of the target class needed to make an accurate prediction, i.e., is the subject suffering from a disease or are they healthy? In other words, the system learns to filter the input information so that only the information within the neural network relevant to the classification task is preserved. MicrobiomePredict is designed to be scalable and can accommodate different input data modalities. During testing, MicrobiomePredict can also manage missing data modalities and still provide accurate predictions.
MicrobiomePredict Data Prediction Pipeline
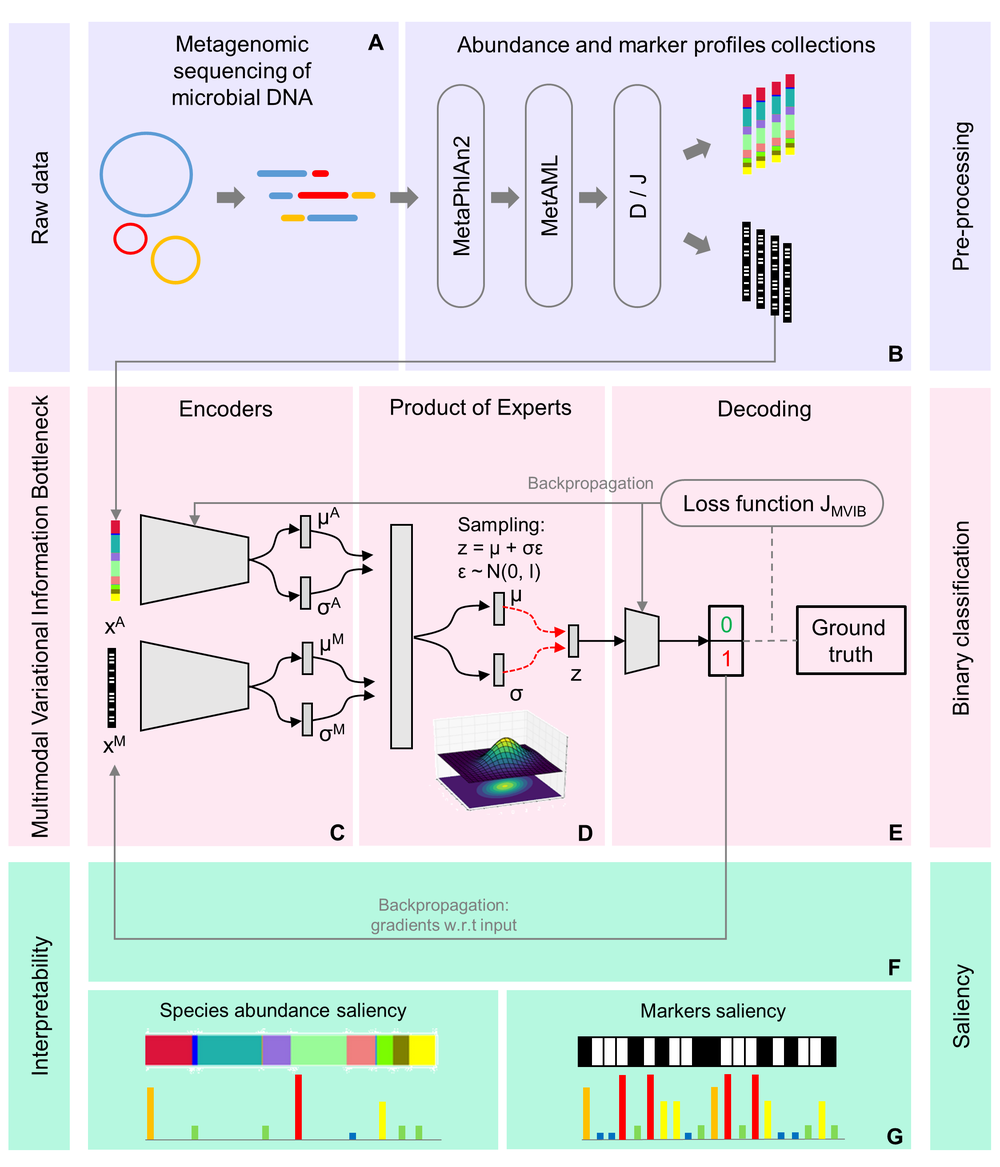
Figure 1: Full workflow. (A, B) data pre-processing. (C, D, E) MVIB. (F, G) explaining predictions with saliency.
Discovering microbiome insights and diagnosing disease with MicrobiomePredict
NEC researchers evaluated MicrobiomePredict on human gut, metagenomic samples from 11 publicly available disease cohorts from six different diseases: Colorectal cancer, obesity, inflammatory bowel disease (IBD), type 2 diabetes, hypertension and cirrhosis of the liver. MicrobiomePredict achieved a high performance (0.80 < ROC AUC < 0.95) with four diseases: colorectal cancer, type 2 diabetes, cirrhosis and obesity.
To achieve interpretability, NEC researchers implemented a method to compute saliency that detect input vector areas that influence predictions of MicrobiomePredict the most. As depicted in Figure 2, the most effective type of gut microbe are significantly more abundant in either healthy (red) or affected individuals (blue). This suggests that MicrobiomePredict is not only an accurate predictor, but can also identify most influential microbiome that causes human disease.
Microbial Species Sorted by Saliency for Colorectal Cancer
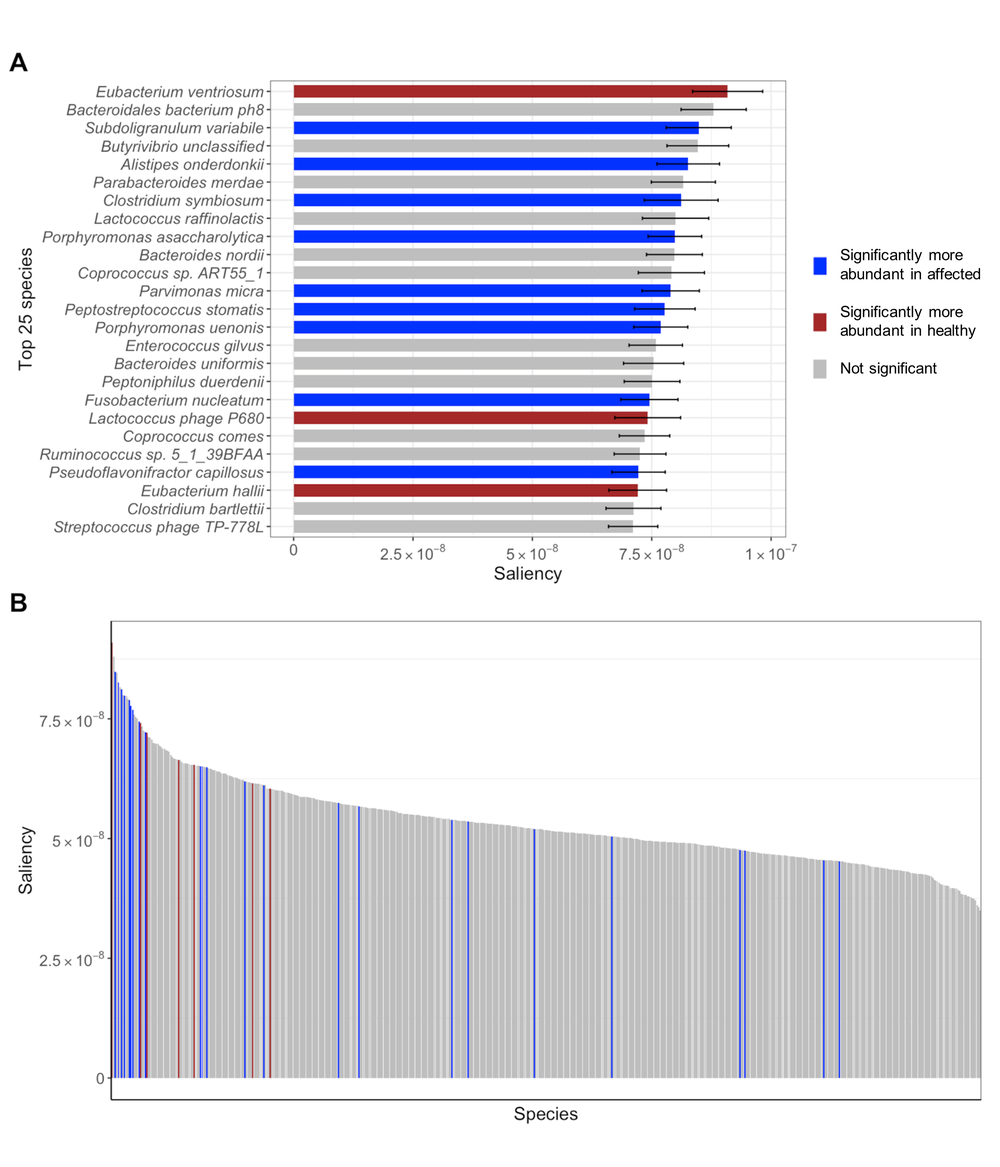
Figure 2: Top 25 microbial species sorted by mean saliency. Relative species-abundance significance in healthy (red) and affected (blue) individuals was calculated using a Wilcoxon signed-rank test for each microbial species for two unpaired samples: healthy and affected individuals.
Many forms of cancer and chronic, lifestyle-related diseases are difficult to predict with any degree of certainty. While much research is focused on improving existing treatment regimens, like immunotherapy, there has been little progress in easily identifying disease prior to or soon after their onset. NEC MicrobiomePredict fills this gap.
The research about MicrobiomePredict was published in PLOS Computational Biology, “Microbiome-based disease prediction with multimodal variational information bottlenecks (F.Grazioli et al.)” and can also be read on our website.